Rules¶
Please read this page carefully, as all participating teams are responsible for adhering to the below rules in order to be considered for placement. The organizers reserve the right to disqualify any participating team found to be in violation of these rules.
As of January 15, 2019, the rules will be considered complete. From that point on, any changes to the rules will be accompanied by an announcement on the forum as well as the announcements page.
Submission Deadline¶
The deadline for submitting test predictions is 23:59 UTC, July 29, 2019. After this time, the submission platform will close until the results are announced, at which time it will reopen and remain open indefinitely.
Submission Format¶
Submissions consist of two parts:
- Test set predictions
- A manuscript describing your methods
1. Predictions¶
Predictions should be submitted as a zip (predictions.zip) archive of
.nii.gz files named
prediction_00210.nii.gz, ..., prediction_00299.nii.gz corresponding to
the ordered list of 90 test cases. That is, you should generate each of
these prediction files in a particular directory, and then from that
directory, run the following command:
zip predictions.zip prediction_*.nii.gz
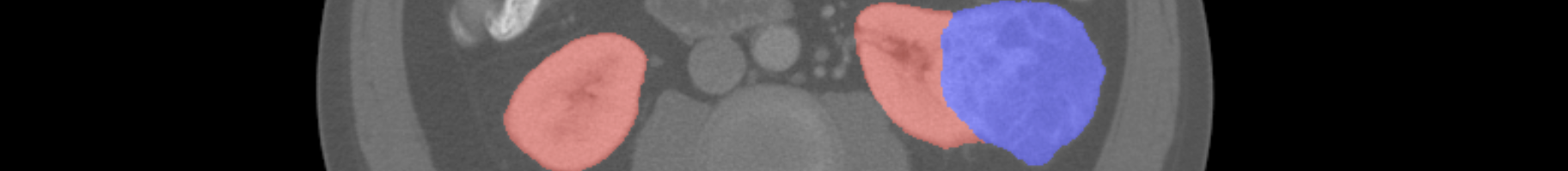
Just like the training labels, values of 0 will be taken to mean background, 1 represents kidney, and 2 represents tumor. Cases missing from the zip archive will be considered entirely background. Values outside of 0, 1, and 2 will also be considered background.
2. Manuscript¶
The submission platform will prompt you for a "Manuscript File" when you submit. You must provide a PDF file here of a manuscript with sufficient detail for a third party to replicate your methods and reproduce your results. You are strongly encouraged to include a link to your source code within your manuscript for this purpose.
There is no strict minimum to manuscript length, but > 4 pages in LNCS format is preferred. You are welcome to submit your manuscript as a technical report, to a preprinting server (e.g. arxiv.org), or even to a fully peer-reviewed venue, but you must provide this file at the time of submission.
Data Use¶
The data was released on March 15, 2019 and will be again on July 15, 2019 under the Creative Commons CC-BY-NC-SA license. Usage of the data must be in accordance with the provisions of that license.
External Data¶
External data may be used, so long as that data is publicly available. This is to allow for the objective and fair comparison of methods. External data use must be described in detail in each team's accompanying manuscript.
Manual Predictions¶
Manual intervention in the predictions of your described automated method is strictly prohibited.
Evaluation and Rankings¶
Teams will be awarded a score for each of the 90 test cases equal to the (Kidney Sørensen–Dice + Tumor Sørensen–Dice)/2. Teams will be ranked by their average score per case, with the highest being the winner. More formally, a team's score, S, will be computed as follows

Where the ith test case has the following confusion matrices
| Predicted Kidney or Tumor | Predicted Background | |
| Labeled Kidney or Tumor |  |
 |
| Labeled Background | 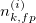 |
| Predicted Tumor | Predicted Not Tumor | |
| Labeled Tumor |  |
 |
| Labeled Not Tumor |  |
A collection of other metrics for both the kidney and tumor segmentation tasks will also be computed and listed on the leaderboard, but they will not affect the ranking. In the unlikely event that two teams achieve the same score up to three significant digits, Tumor Sørensen–Dice will be used as a tie-breaker.